
Figure 1. Found throughout the United States but especially in warm and coastal areas, take-all root rot is a major infectious disease of St. Augustinegrass caused by the soil-borne fungus species of Gaeumannomyces. Photos courtesy of the lab of Young-Ki Jo
Maintaining healthy grasses at a high level of aesthetic quality and playability is quite a daunting task, as numerous pests and diseases threaten to diminish that quality. Our lack of understanding of causal agents leads to generalizations in management being made that may prove to be less effective in treating and controlling diseases. Such has been the case with the fungal disease take-all root rot in warm-season turfgrasses.
Found throughout the United States but especially in warm and coastal areas, take-all root rot is a major infectious disease of St. Augustinegrass (Figure 1), which is caused by the soil-borne fungus species of Gaeumannomyces. Take-all root rot typically becomes steadily pronounced when yellowing patches appear during stressful periods, such as during hot and dry summer weather. Although roots and stolons may have been infected for some time prior, additional abiotic stresses on the plant give the disease an opportunity to expand. As the disease progresses, infected grasses will start to turn thin and bare, as black and rotting spots on the roots hinder nutrient uptake. These irregular or diffused patches of symptoms can grow anywhere from 1 foot (0.3 meters) to over 20 feet (6 meters) in diameter.
Symptoms in St. Augustinegrass may be mistaken for large patch caused by the fungus Rhizoctonia solani or chinch bug infestations. The key differences from take-all root rot are that large patch infects and rots the plant stems usually in the fall season, and chinch bug is an insect feeding on the plants.
St. Augustinegrass is not alone in its susceptibility to take-all root rot. Various species in the Gaeumannomyces genus are responsible for infecting various grasses — including bermudagrass, centipedegrass and zoysiagrass — with root-decline diseases. In addition, Gaeumannomyces species are attributed to be the cause of root rotting in cool-season turfgrasses and monocot crops, including rice, wheat and oats (5).
With such wide ranges in variation among Gaeumannomyces species, the differences are mostly attributed to primary plant hosts, morphology of cultured fungi, and genetic variation, the study of which culminated in 2016 with a reclassification scheme of Gaeumannomyces fungi (3). Previously, the causal fungus of take-all root rot was Gaeumannomyces graminis var. graminis. However, multiple organisms once grouped together as Gaeumannomyces graminis var. graminis were divided into their own separate species, such as G. australiensis (which infects wheat), G. fusiformis (which infects rice), and G. graminis (which infects bermudagrass). Four of the newly classified species from Gaeumannomyces graminis var. graminis infect St. Augustinegrass — G. arxii, G. californicus, G. floridanus and G. graminicola. However, the distribution of these species, their unique characteristics and their relative pathogenicity (or severity of infection) remained unknown.
In light of these developments, we began a reexamination of take-all root rot and its causal Gaeumannomyces fungi as it affects St. Augustinegrass. Our primary goal was to better understand the diversity of Gaeumannomyces fungi commonly found in St. Augustinegrass in Texas as well as the severity by which they can inhibit plant performance.
Classifying take-all root rot pathogens
Seventy-five samples of various Gaeumannomyces species were isolated from St. Augustinegrass in 21 Texas counties. Stolons with ectotrophic hyphae and lobed hyphopodia (Figure 2, blackened areas with webbed fungal mycelia on the plant surface) were cut and washed into 2-inch (5.1-centimeter) or smaller pieces, then sterilized with bleach and rinsed.
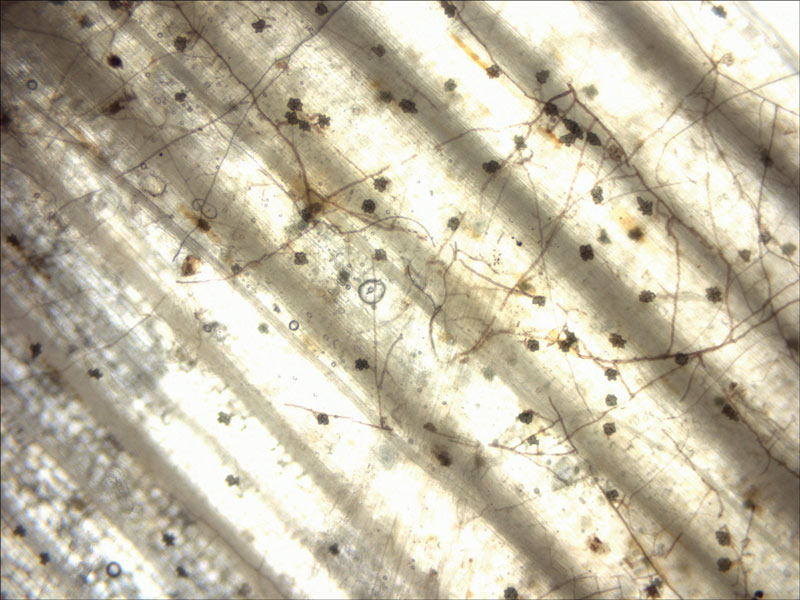
Figure 2. A microscopic image of the soil-borne fungus species of Gaeumannomyces on the plant surface. Dark hyphae produce hyphopodia (flower-like lobed outgrowths), which serve to attach to and infect the plant.
Once cleaned, the samples were placed into petri dishes containing potato dextrose agar (PDA) amended with fungicides (mefenoxam, flutolanil and iprodione) and an antibiotic (streptomycin sulfate) to create a nutrient base that allows the growth of Gaeumannomyces species but halts the growth of other undesirable bacteria and fungi. Once plated, these petri dishes were kept in a dark incubator at 77 F (25 C) for several days. Fungal colonies exhibiting characteristics of Gaeumannomyces species were then isolated.
Mycelial formation and appearance of Gaeumannomyces species on PDA was the first step in identifying and sorting the samples. Gaeumannomyces species are slow-growing, often requiring two weeks or more before reaching their maximum size and form. Colony shape (round or irregular), color (degree of melanization or darkening), growth rate (colony expansion on the plate per day), and hyphae structure (lobed hyphopodia, perithecia, phialidic structures) are some of the components used to visually differentiate samples. Measuring the growth rate under various temperatures (68, 77, 86 and 95 F; 20, 25, 30 and 35 C) was another way to sort isolates into their proper species.
In addition to the phenotypes of the samples, their genetics were also used as a source of identification. We used genetic differences in specific indicator regions of three genes — internal transcribed spacer region (ITS), partial 28S rDNA and partial RNA polymerase II largest subunit gene (rpb1). Mycelium of each fungal isolate was collected, and its DNA was extracted and purified. Using polymerase chain reactions (PCR), these indicator genes of Gaeumannomyces isolates were amplified. These amplified genes were sequenced at the sequencing facility. Once done, they were checked and compared against previously published sequences as maintained in the NCBI GenBank Database, a major repository for genetic sequences.
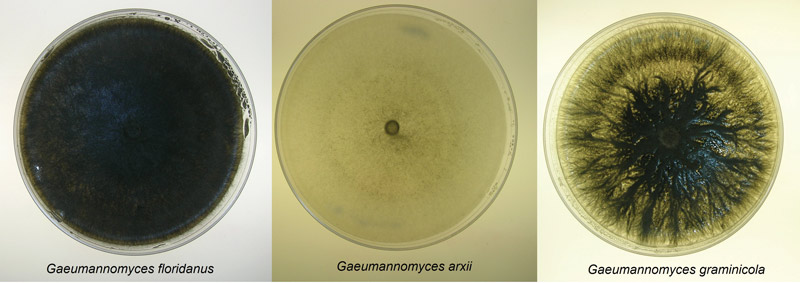
Figure 3. Culture plates of three of the newly classified species of Gaeumannomyces that infect St. Augustinegrass.

Figure 4. A side-by-side comparison illustrates the difference between inoculated and uninoculated plants.
Three distinct morphological groups on PDA were found, each correlating with a different species (Figure 3). Dark pigmentation with round colony formations were matched with G. floridanus; those with round colonies but little coloring were attributed to G. arxii; and irregular colony shapes with dark coloring indicated G. graminicola. Isolates grew best in 77 F to 86 F, and G. arxii failed to grow at 95 F (Figure 4). Genetic sequences for each species used in this study matched those described in Hernandez-Restrepo et al. (3), further confirming the distinctions.
Pathogenicity of take-all root rot pathogens
With take-all root rot fungal samples identified, the next step was to understand their pathogenicity — that is, how severely they can infect plants. Given the difficulty of finding St. Augustinegrass plants not already infected with some form of take-all root rot fungi and similar interactions of Gaeumannomyces with St. Augustinegrass and rice (1, 2), rice seedlings (Presidio variety) were selected as alternative host plants to test the pathogenicity of take-all root rot fungi.
Cone-tainers were filled with vermiculite until 1.25 inches (3.18 centimeters) from the top. Five plugs of actively growing mycelium were placed at this level, then covered with additional vermiculite until the cone-tainer was filled. Cleaned rice seeds were then planted in them and placed in a growth chamber for 26 and 42 days. Once the time was up, the plants were removed, their roots carefully washed, shoot height and root length measured, and root lesion severity assessed.
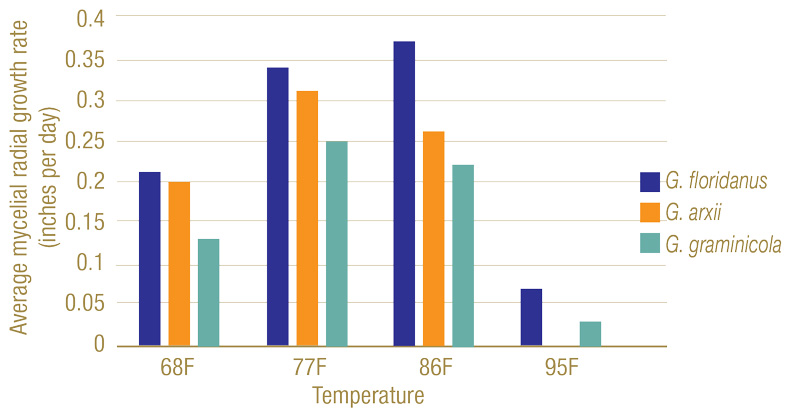
Figure 5. Mycelial radial growth rates of Gaeumannomyces species on potato dextrose agar at varying temperatures. While 77 F to 86 F (25 C to 30 C) proved ideal for growth, all had a significant decline at 95 F (35 C), with G. arxii samples having no growth.
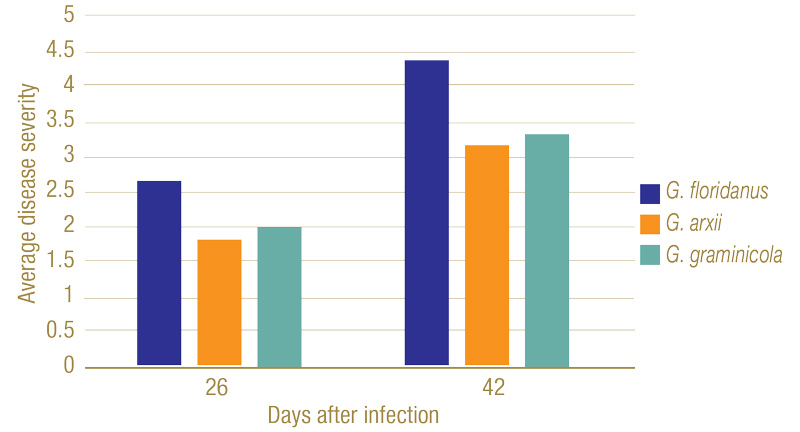
Figure 6. Disease severity ranking of plant roots 26 and 42 days after infection. On the ranking scale developed by Datnoff et al., 1997, 1 = no discoloration of the root system; 2 = 1 to 25% of roots exhibiting black lesions or general tan discoloration; 3 = 26 to 50% of roots with black coalescing lesions; 4 = 51 to 75% of roots with black coalescing lesions; and 5 = 76 to 100% of roots with black lesions.
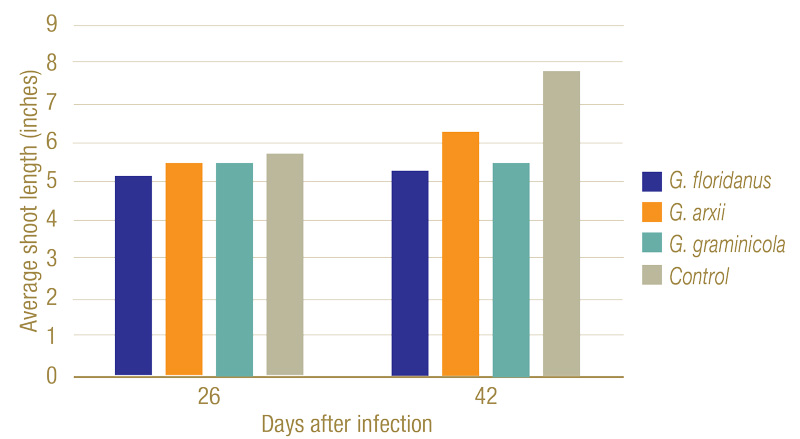
Figure 7. Average shoot lengths of plants 26 and 42 days after infection. While slightly shorter, no statistically significant changes in height could be seen after 26 days. By 42 days, however, the fungi had a clear negative impact on the plant’s growth.
Inoculated Gaeumannomyces species produced dark lesions on stems and roots of rice seedlings (Figure 5). After 26 days, G. floridanus had the highest average disease severity rating, followed by G. arxii and G. graminicola (Figure 6). This trend of greater aggression by G. floridanus was also true after 46 days. After 42 days of infection, Gaeumannomyces species did show lower shoot heights than the uninoculated control (Figure 7). G. arxii tended to have the tallest shoots of the inoculated plants, but they were still on average 1.5 inches (3.8 centimeters) shorter than the healthy controls. Root lengths proved to be similar across all samples.
Discussion
Prior to 2016, Gaeumannomyces species were commonly thought to be very similar, hence their classification as a single species of Gaeumannomyces graminis var. graminis. Our study shows the real complexity behind Gaeumannomyces species and why a reexamination differentiating them is important for both scientists and turfgrass managers.
While they all are found in St. Augustinegrass, the unique characteristics highlighted between G. floridanus, G. arxii and G. graminicola show that levels of effects can be quite different. Confirming such differences, both genetically and phenotypically, is a key step to managing take-all root rot properly.
Many general management practices for turfgrass may take measures to prevent take-all root rot from appearing in their areas. Reducing thatch, proper drainage, maintaining proper mowing heights and balancing soil pH are known ways to prevent the disease from manifesting. Use of fungicides to prevent outbreaks is a necessary method for high-end turfgrass systems such as golf courses, in particular with azoxystrobin, myclobutanil, propiconazole and thiophanate-methyl being among the most commonly used. All these steps, however, are generalized. With the improved diagnostics, we can develop more effective and localized treatments of take-all root rot.
Understanding field conditions and how they interact with take-all root rot and plant health are critical to improving management. Of note in our research, G. floridanus was generally more aggressive than G. arxii or G. graminicola. The fact that G. floridanus has greater survivability in warmer temperatures compared with the others also suggests a greater adaptability to subtropical and tropical regions where St. Augustinegrass is most popularly cultivated, indicating a greater potential problem for management and a need for more vigorous management. Yet the frequent appearance of the more benign G. arxii and lack of correlation between species and geographic location where the samples originated means that determining the exact species will be an important first step for such specific controls.
Such can be seen in take-all root rot research conducted on golf course bermudagrasses (4). While showing similar high variance between fungal samples and optimal fungal growth in 76 F to 82 F, G. graminicola and other Gaeumannomyces species were shown to be the most aggressive compared with two other related fungi that were only moderately aggressive. This means that not only should environment and fungal species be considered when developing management plans, but the host grass as well.
With this foundation firmly reestablished, the next phase to combat take-all root rot can begin, one in which management practices are precisely and pragmatically targeted. Research focusing on not only learning more on the variance in pathogenicity but also potential fungicide sensitivities is already underway.
Funding
This study was supported by the USGA (2020-16-721).
Acknowledgements
We thank the Texas Plant Disease Diagnostic Lab for providing turfgrass samples used in this study and Raleigh Darnell for assisting with lab assays. The most data presented in this article was published in: Zidek, Matthew J., Lin Yu, Michael Jochum and Young-Ki Jo. 2021. Complexity of Gaeumannomyces species causing take-all root rot of St. Augustinegrass in Texas. Mycologia 113(3): 599-611 (https://doi.org/10.1080/00275514.2021.1881735).
The research says ...
- Take-all root rot fungi are ubiquitously found in warm-season turfgrasses.
- Environmental factors stressing turfgrass as well as take-all root rot infection attribute to disease symptom development.
- The presence of the take-all root rot fungi does not necessarily lead to plant death and turf damage but compromises plant health.
- Identification of Gaeumannomyces species can help prepare better turfgrass management practices.
Literature cited
- Datnoff, L.E., M.L. Elliott and J.P. Krausz. 1997. Cross pathogenicity of Gaeumannomyces graminis var. graminis from bermudagrass, St. Augustinegrass, and rice in Florida and Texas. Plant Diseases 81(10):1127-1131 (https://doi.org/10.1094/PDIS.1997.81.10.1127).
- Dufresne, M., and A.E. Osbourn. 2001. Definition of tissue-specific and general requirements for plant infection in a phytopathogenic fungus. Molecular Plant-Microbe Interactions 14(3):300-307 (https://doi.org/10.1094/MPMI.2001.14.3.300).
- Hernandez-Restrepo, M., J.Z. Groenewald, M.L. Elliott, G. Canning, V.E. McMillan and P.W. Crous. 2016. Take-all or nothing. Studies in Mycology 83:19-48 (https://doi.org/10.1016/j.simyco.2016.06.002).
- Stephens, C., T.W. Gannon, M. Cubeta, T.L. Sit and J. Kerns. 2021. Characterization and aggressiveness of take-all root rot pathogens isolated from symptomatic bermudagrass putting greens. Phytopathology 1-40 (https://doi.org/10.1094/PHYTO-05-21-0215-R).
- Walker, J. 1981. Taxonomy of take-all fungi and related genera and species. In: M. Asher and P. Shipton (editors). Biology and control of take-all. London, United Kingdom: Academic Press.
Paul Goetze is an Extension assistant, and Young-Ki Jo is a professor and Extension specialist in the Department of Plant Pathology and Microbiology at Texas A&M University and Texas A&M AgriLife Extension Service.